Provisional PDF - Parasites & Vectors PDF
Preview Provisional PDF - Parasites & Vectors
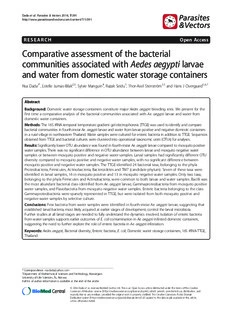
Dadaetal.Parasites&Vectors2014,7:391 http://www.parasitesandvectors.com/content/7/1/391 RESEARCH Open Access Comparative assessment of the bacterial communities associated with Aedes aegypti larvae and water from domestic water storage containers Nsa Dada1*, Estelle Jumas-Bilak2,3, Sylvie Manguin4, Razak Seidu1, Thor-Axel Stenström1,5 and Hans J Overgaard1,6,7 Abstract Background: Domestic water storage containers constitute major Aedes aegypti breeding sites. We present for the first time a comparative analysis ofthe bacterial communities associated with Ae. aegypti larvae and water from domestic water containers. Methods: The16SrRNA-temporaltemperaturegradientgelelectrophoresis(TTGE)wasusedtoidentifyandcompare bacterialcommunitiesinfourth-instarAe.aegyptilarvaeandwaterfromlarvaepositiveandnegativedomesticcontainers inaruralvillageinnortheasternThailand.WatersampleswereculturedforentericbacteriainadditiontoTTGE.Sequences obtainedfromTTGEandbacterialcultureswereclusteredintooperationaltaxonomicunits(OTUs)foranalyses. Results:SignificantlylowerOTUabundancewasfoundinfourth-instarAe.aegyptilarvaecomparedtomosquitopositive watersamples.TherewasnosignificantdifferenceinOTUabundancebetweenlarvaeandmosquitonegativewater samplesorbetweenmosquitopositiveandnegativewatersamples.LarvalsampleshadsignificantlydifferentOTU diversitycomparedtomosquitopositiveandnegativewatersamples,withnosignificantdifferencebetween mosquitopositiveandnegativewatersamples.TheTTGEidentified24bacterialtaxa,belongingtothephyla Proteobacteria,Firmicutes,Actinobacteria,BacteroidetesandTM7(candidatephylum).Sevenofthesetaxawere identifiedinlarvalsamples,16inmosquitopositiveand13inmosquitonegativewatersamples.Onlytwotaxa, belongingtothephylaFirmicutesandActinobacteria,werecommontobothlarvaeandwatersamples.Bacilliwas themostabundantbacterialclassidentifiedfromAe.aegyptilarvae,Gammaproteobacteriafrommosquitopositive watersamples,andFlavobacteriafrommosquitonegativewatersamples.Entericbacteriabelongingtotheclass GammaproteobacteriaweresparselyrepresentedinTTGE,butwereisolatedfrombothmosquitopositiveand negativewatersamplesbyselectiveculture. Conclusions:Fewbacteriafromwatersampleswereidentifiedinfourth-instarAe.aegyptilarvae,suggestingthat establishedlarvalbacteria,mostlikelyacquiredatearlierstagesofdevelopment,controlthelarvalmicrobiota. Furtherstudiesatalllarvalstagesareneededtofullyunderstandthedynamicsinvolved.Isolationofentericbacteria fromwatersamplessupportsearlieroutcomesofE.colicontaminationinAe.aegyptiinfesteddomesticcontainers, suggestingtheneedtofurtherexploretheroleofentericbacteriainAe.aegyptiinfestation. Keywords:Aedesaegypti,Bacterialdiversity,Entericbacteria,E.coli,Domesticwaterstoragecontainers,16SrRNA-TTGE, Thailand *Correspondence:[email protected] 1DepartmentofMathematicalSciencesandTechnology,Norwegian UniversityofLifeSciences,Ås,Norway Fulllistofauthorinformationisavailableattheendofthearticle ©2014Dadaetal.;licenseeBioMedCentralLtd.ThisisanOpenAccessarticledistributedunderthetermsoftheCreative CommonsAttributionLicense(http://creativecommons.org/licenses/by/4.0),whichpermitsunrestricteduse,distribution,and reproductioninanymedium,providedtheoriginalworkisproperlycredited.TheCreativeCommonsPublicDomain Dedicationwaiver(http://creativecommons.org/publicdomain/zero/1.0/)appliestothedatamadeavailableinthisarticle, unlessotherwisestated. Dadaetal.Parasites&Vectors2014,7:391 Page2of12 http://www.parasitesandvectors.com/content/7/1/391 Background the mosquitoes, or are transient and lost during digestion Thecontainer-breedingmosquito,Aedesaegypti,isawell- and/or molting [10]. A better understanding of the dy- recognized vector of diseases of significant public health namics involved in bacterial colonization within mosqui- concern, such as dengue fever, yellow fever, and chikun- toesisthereforestillneeded. gunya [1]. Aedes aegypti is known to breed mainly in Furthermore,understandingthemosquito-bacteriainter- domesticwatercontainersinandaroundhumandwellings actions within Ae. aegypti infested domestic containers [2-4]. These containers are ecosystem microcosms sup- would be beneficial for Ae. aegypti control. Thus far, one porting food webs that are dependent on bacteria [5]. study that has examined the bacterial content of Ae. Studies conducted on the microbial ecology of Aedes aegyptiinfesteddomesticcontainers[9]employedconven- breeding containers have mainly focused on the invasive tional culture-dependent methods. Thesemethods are un- Ae. albopictus as well as other tree-hole mosquitoes e.g. abletocapturenon-cultivablebacteriathatmaybepresent Ae. triseriatus and Toxorhynchites rutilus. These studies within samples, thus leading to an incomplete picture of have explored the microbial communities in natural con- the bacterial communities. This contributes to the vague tainers such as tree holes, discarded or unused containers knowledgeor limited information available on the dynam- (e.g.tyres),andornamentalcontainers(e.g.plantpotsand ics involved in the mosquito-bacteria interactions. How- cemetery urns) [5-8]withno focusonthe microbialecol- ever,molecularmethodssuchasthetemporaltemperature ogyofAe.aegyptiinfesteddomesticwatercontainers.One gradient gel electrophoresis (TTGE) and 16S rRNA se- studyontheeffectofAe.aegyptimidgutmicrobiotaonits quencing can identify both cultivable and non-cultivable susceptibility to DENV-2 however characterized the bac- bacteriaalthoughnotwithoutlimitations[18]. terial content of domestic water containers from which Inthisstudy,the16SrRNA-TTGEtechniquewasused themosquitoeswerecollected[9]. to comparatively assess the bacterial communities asso- Mosquito-microbe interactions are of increasing re- ciated with Ae. aegypti larvae and water from domestic search interests [10]. These interactions which span water containers. We hypothesized that the bacteria in from pathogenic to obligate symbiosis[11],usually affect Ae. aegypti infested containers are determinants of Ae. the evolutionary success and physiological functions of aegypti productionandthus may constitute amajor pro- the mosquitoes. Such functions could be beneficial to portion of the larval microbiota. This was based on our the mosquitoes, for example, in the synthesis of essential previousstudywheresignificantlymoreAe.aegyptipupae nutrients that may be lacking from food sources, and con- were produced in domestic containers contaminated with ferral of resistance to pathogens [10,12], or detrimental, Escherichia coli compared to containers that were not where the microbiota directly interfere with mosquito de- contaminatedwithE.coli[19].Totestthishypothesisand velopmentand fitness[10,13]. Theseinteractions arebeing update information on the bacterial communities asso- exploredforthedevelopmentofnovelcontrolstrategiesfor ciated with Ae. aegypti larvae and water in Ae. aegypti mosquitovectorsandmosquito-bornediseases[12],anap- infested domestic containers, the abundance and diversity proachrecentlytermed‘symbioticcontrol’[14]. of bacterial taxa between Ae. aegypti larvae and water Bacteria constitute a proportion of mosquito micro- fromAe.aegyptipositiveandnegativecontainerswerede- biota, colonizing a variety of mosquito organs, chiefly terminedandcompared. the midgut, and to a lesser extent the hemolymph, saliv- aryglandsandreproductiveorgans [10,11].Thebacterial Methods communities within mosquitoes vary depending on the Samplingsitesandsamplecollection mosquito species, sex, stage of development and habitat The study was conducted in February 2012 following our [10]. The origin of bacteria within mosquitoes and the earlier study on the relationship between Ae. aegypti pro- dynamics involved in bacterial colonization are unclear duction and E. coli contamination in domestic containers as both exogenous and endogenous factors are known to in Thailand and Laos [19]. Mosquito and water samples affect the initial colonization and nature of the bacterial werecollectedfromdomesticwatercontainersinWaileum composition [10]. Some studies show that immature village,KhonKaenprovince,Thailand,where25outof122 mosquitoes or newly emerged adults harbor bacteria de- houses included in the previous study were randomly se- rived from their breeding habitats [10,15]. Others show lected.Outoftherandomlyselectedhouses,17hadatleast that the bacterial communities in blood feeding adults one mosquito positive container, and were included in this are influenced by their blood meals [16]. Feeding may, study. The other eight houses were excluded because they thus, play a role in determining bacterial communities were either unoccupied at the time of sampling or had no within mosquitoes. The bacteria within mosquitoes have mosquitopositivecontainers. alsobeenshowntobeacquiredtransstadially[17].Yetitis In each house, water samples were collected from still uncertain whether these bacteria acquired transsta- each mosquito positive container and one mosquito dially or through feeding are able to survive and colonize negative container directly into 100 ml standard Dadaetal.Parasites&Vectors2014,7:391 Page3of12 http://www.parasitesandvectors.com/content/7/1/391 Whirl-Pak® Thio-Bags®. Water was vigorously mixed on the other half of each filter membrane using prior to sampling to ensure that biofilms were in- the Master Pure Gram Positive DNA purification kit® cluded.Atotalof25Ae.aegyptipositivecontainers(10 (Epicentre Biotechnologies, Madison, USA) following cement tanks, 12 earthen jars, two plastic buckets and manufacturer’s instructions. Each sample was disrupted one plastic drum), and 17 negative containers (13 in 150 μl Tris-EDTA (TE) buffer solution with the aid of earthen jars, two cement tanks, one plastic drum and sterile plasticpestlesbefore DNAextraction. one metal pot) were sampled. A sample of ten 4th in- DNA from pure bacterial colonies was extracted by star larvae (or all if less than ten) was collected from boiling. Cryopreserved pure bacterial colonies were re- each mosquito positive container and transferred to grown on TSA at 37±0.5°C for 24 hours prior to DNA sterile 15 ml Eppendorf tubes. All water and larval extraction. Three to six colonies, depending on size, samples were transported on dry ice to the laboratory were picked from TSA plates and mixed with 100 μl of andstoredin-80°Cuntilfurtherprocessing. DNAfreewaterinsterile1.7mlEppendorftubes.Thecell suspensions were held in a boiling water-bath for 10 min SamplepreparationforDNAextraction to lyse the cells, then vigorously homogenized withavor- Samples were thawed at room temperature before DNA tex mixer for 10 sec and chilled on ice. Resulting DNA extraction. Aedes aegypti larvae were identified using il- sampleswerestoredin-20°CforPCR. lustrated keys [20], and all Ae. aegypti larvae (mean 4± 1) per container were pooled together for analysis. A PCRamplificationofbacterialsmallsubunitrRNAgene total of 25 pools of Ae. aegypti larvae from 25 mosquito andTTGEanalysis positivecontainerswereanalyzed.Larvalsamplesweresur- Bacterial DNA from mosquito larvae and water samples face sterilized; first rinsed in 70% ethanol, then suspended were used as PCR templates. For mosquito and uncul- in 70% ethanol and agitated with a vortex mixer for about tured water samples, theV2-V3 region of the 16S rRNA 10seconds,andfinallyrinsedwithsterileDNAfreewater. gene was amplified using universal 16S rRNA bacterial Each water sample was filtered through a cellulose ni- primers HDA1: 5′-ACTC CTA CGG GAG GCA GCA trate membrane filter (0.45 μm pore size, 47 mm dia., GT-3′, and HDA2: 5′-GTATTA CCG CGG CTG CTG Sartorius Stedium®) using aseptic vacuum filter units GCA-3′,which yieldPCRfragmentsof~199bp.The16S (Millipore®). 0.45 μm pore size membrane filters were rRNA gene from cultured water samples was amplified used due to high turbidity of some water samples. Each using the primers 27f: 5′-GTGCTGCAGAGAGTTTG membrane filter was cut in two halves; one half cut into ATCCTGGCTCAG-3′), and 1492r: 5′-CACGGATCCT small piecesandplaced insterile 1.5ml Eppendorftubes ACGGGTACCTTGTTACGACTT-3′, which yield PCR for DNA extraction and the other placed on Trypticase fragmentsof~1469bp.Amplificationwascarriedoutfol- soy agar (TSA) plates for bacterial cultures and further lowing previously reported protocols [21] and [18] re- isolation of enteric bacteria. Prior to water sample filtra- spectively. PCR products obtained from mosquito larvae tion, sterile DNA free water was filtered through each and uncultured water samples were separated by TTGE. filter unit (using separate membranes) as negative con- TTGEmigrationwasperformedfollowingapreviouslyre- trols tocheckfor contamination. portedprotocol[21]on16cm×16cm×1mmgelsusing the DCode universal mutation detection system (Bio-Rad Isolationofentericbacteriafromwatersamples Laboratories, Marne-la-Coquette, France). PCR prod- One half of each filter membrane placed on TSA plates ucts from pure bacterial cultures and about 50% of the were incubated at 37±0.5°Cfor 48hours.Colonies from bandsproducedbyTTGEweresequenced.Theremaining positive TSA plates were cryopreserved in Tryptic soy bands were assigned to an Operational Taxonomic Unit broth (TSB) with 15% glycerol at -80°C for isolation of (OTU) by comparing their migration distance to that of enteric bacteria. Enteric bacteria were isolated from pre- sequencedbands. served samples by streaking on Drigalski agar (DA) plates. Following incubation at 37±0.5°C for 48 hours, Sequenceanalysis,taxonomyassignmentandalignment each colony displaying distinct morphotypes on positive The sequences were analyzed using the Quantitative In- DA plates were isolated and sub-cultured on fresh DA sights Into Microbial Ecology (QIIME) software package platestoobtainpuresingle colonies.Puresingle colonies version 1.7.0 [22]. Bacterial diversity of a sample ana- were cryopreserved in TSB with 15% glycerol at -80°C lyzed using TTGE is generally reflected by the number forsubsequent DNA extraction. of bands on the TTGE profile, each band usually repre- senting an OTU. However, Manguin et al. [21] reported GenomicDNAextraction bands with different migration distances belonging to the Bacterial genomic DNA was extracted from pools of sameOTUduetosequenceheterogeneityamongtheir16S sterilized Ae. aegypti larvae and bacterial cells retained rRNAgene copies.Thiscouldleadtoanoverestimationof Dadaetal.Parasites&Vectors2014,7:391 Page4of12 http://www.parasitesandvectors.com/content/7/1/391 the bacterial diversity. Therefore to avoid this, sequences in boxplots. Prior to diversity analyses samples were rar- obtained from TTGE aswell asthoseobtained from water efied down to the mean number of sequences per sam- samplecultureswereclusteredintoOTUs,priortoanalysis. ple – four for samples analyzed by TTGE, and two for Sequences were clustered into OTUs using the UCLUST water sample cultures – in ten iterations to standardize pipeline[23] in QIIME at anidentitythreshold of 97% (i.e. number of sequence per sample. Level of statistical sig- sequencesthatwere97%similarwerebinnedintothesame nificanceforallanalyseswassetat p≤0.05. OTU). The most abundant sequence within each cluster wasselected as a representative of the OTU. The resulting Results OTUswereassignedtotaxausingtheRibosomalDatabase Datasummary Project(RDP-II)classifier[24]trainedonGreengenesrefer- TTGE profiles were obtained from 18 (72%) pooled ence database [25] via QIIME at a minimum confidence mosquito samples, 22 (88%) mosquito positive and 16 score of 80%. The OTUs were aligned against the Green- (80%) mosquito negative water samples. The remaining genescorereferencealignment[26]usingthePythonNear- samples showed no PCR amplification or faint PCR sig- est Alignment Space Termination (PyNAST) alignment nals leading to undetectable TTGE profiles. A total of algorithm [27] in QIIME with a minimum identity of 75%. 236 sequences were generated from these TTGE bands; Relativeabundanceofbacterialtaxawascomputedandcol- 61frommosquitolarvae,117and58frommosquitoposi- lated using the make_OTU_table.py and summarize_taxa. tive and negative water samples respectively. The se- pyscriptsinQIIME.Thiswasvisualizedonhistogramsand quences were binned into 134 unique OTUs, with 1-14 comparedusingtwo-samplet-testsinExcel.Aphylogenetic sequences per OTU, and average sequence length of tree, required to run diversity analyses down the QIIME 120 bp. Seven of the unique OTUs failed to align to any pipeline,wasconstructedusingtheFastTreeapproximately bacterial small subunit models (SSU) and were discarded. maximum likelihood program [28] in QIIME. Representa- Sequencesfrom12poolsofmosquitos,11mosquitoposi- tiveOTUsequencesgeneratedfromthisstudyareavailable tivecontainers,and10mosquitonegativecontainerswere in the GenBank database [GenBank:KJ814977 - KJ815004; includedindiversityanalysesafterrarefaction. KM108474-KM108577]. Water samples from all mosquito negative containers (17) and 23 (92%) mosquito positive containers produced Diversityanalysis bacterial colonies on selective media for enteric bacteria. Diversity analysis was performed separately on bacterial A total of 111 bacterial sequences were obtained from sequences obtained by TTGE (mosquito and uncultured these colonies; 69 from mosquito positive containers and water samples), and on those obtained from water sam- 42 from mosquito negative containers. These sequences ples cultures. The OTU diversity within and between were binned into 24 unique OTUs, with 1-5 sequences samples were compared, using alpha (α) and beta (β) di- perOTU,andaveragesequencelengthof911bp.Sevenof versity indices respectively. Alpha diversity was mea- the OTUs failed to alignto any bacterialSSU models and sured with the Shannon-Wiener index (relative OTU werediscarded.Sequencesfrom19mosquitopositive,and abundance and evenness), Observed species metrics 13 mosquito negative water samples were included in di- (OTU abundance), and Faith’s whole tree Phylogenetic versityanalysesafterrarefaction. Diversity (branch length-based diversity) [29]. Alpha di- versity means and two-sample t-tests were computed AlphadiversityofbacterialOTUs using Excel. Beta diversity was evaluated using the un- Figure 1 shows alpha diversity rarefaction curves based weighted UniFrac [30] pipeline in QIIME. Principal Co- ontheShannonWienerindex,observedspeciesindexand ordinate Analysis (PCoA) was used to interpret and Faith’s phylogenetic diversity for sequences generated by visualize the variation in UniFrac distance matrix. The TTGE. Mosquito positive water samples had the highest largest amount of variation is explained by the first prin- OTU abundance, while mosquito larvae had the lowest cipal coordinate (PCo1), the second largest by the sec- across all the three indices (Figure 1). Two-sample t-tests ond principal coordinates (PCo2) and so on. To test the comparing each index between samples revealed statis- strength and significance of the PCoA, the adonis func- tically significant results for only the Shannon-Wiener tioninPRIMER6(PRIMER-ELtd.,Luton,UK)wasused index, which was significantly higherinmosquitopositive on unweighted UniFrac distance matrices via QIIME. water samples compared to mosquito larvae (p<0.05). The adonis function computes an effect size value R2, The difference was not statistically significant between which shows the percentage of variation in distance mosquito larvae and mosquito negative water samples, or matrices explained by the sample. Two-sample student’s betweenmosquitopositiveandnegativewatersamples. t-test to compare mean unweighted UniFrac distances For sequences obtained from bacterial cultures of water between samples was calculated using the dissimilari- samples,OTUabundancewashigherinmosquitopositive ty_mtx_stats.py script in QIIME with results presented containers compared to mosquito negative containers Dadaetal.Parasites&Vectors2014,7:391 Page5of12 http://www.parasitesandvectors.com/content/7/1/391 A Shannon-Wiener index 1.4 x 1.2 e d n 1 I n o 0.8 n n a 0.6 h S 0.4 0.2 0 1 2 3 4 7 B Observed species index 6 e c5 n a nd4 u b a3 U T2 O 1 0 1 2 3 4 C Faith's phylogenetic diversity 1.8 1.6 y sit 1.4 r ve 1.2 c di 1 eti 0.8 n e g 0.6 o yl 0.4 h P 0.2 0 1 2 3 4 N (sequence/sample) Ae. aegypti larvae Mosquito negative water samples Mosquito positive water samples Figure1AlphadiversityrarefactioncurvesofbacterialOTUsfromTTGE.AlphadiversityrarefactioncurvesofbacterialOTUsfromAe.aegypti larvae,mosquitopositiveandmosquitonegativewatersamplesanalyzedbyTTGE.ThisisbasedonShannon-Wienerindex(A),Observedspeciesindex (B)andFaith’sphylogeneticdiversity(C).Curvesrepresentmeandiversityindicesforeachsample,anderrorbarsrepresentstandarderrorofmeans. acrossallthreeindices,butthedifferencewasnot statisti- variation in UniFrac distances, and revealed distinct clus- callysignificant(p>0.05). teringpatternsofAe.aegyptilarvaeandmosquitopositive water samples (Figure3). ThePCoA plot shows a distinct Betadiversity cluster of Ae. aegypti larvae towards the right of the first The unweighted UniFrac distance metrics was used to plot between PCo1 and PCo2, as well as the second plot compare OTU diversity between samples. TheTTGE re- between PCo1 and PCo3. This cluster is also visible to- sults showed significantly different and lower bacterial wards the left of the third plot between PCo2 and PCo3 diversityinAe.aegyptilarvaecomparedtomosquitoposi- (Figure 3). On all three PCoA plots, mosquito positive tive (p<0.0001) and negative (p<0.0001) water samples water samples cluster towards the bottom left, separately (Figure2). Therewas nosignificant difference in bacterial from mosquito larvae (Figure 3). The mosquito negative diversity between mosquito positive and negative water water samples seem to spread out over all three plots samples (p>0.05). The PCoA captured over 44% of withoutanydistinctclustering(Figure3).Theadonistest, Dadaetal.Parasites&Vectors2014,7:391 Page6of12 http://www.parasitesandvectors.com/content/7/1/391 Figure2BoxplotsofunweightedUniFracdistancesofbacterialOTUsfromTTGE.BoxplotsshowdistributionofunweightedUnifrac distancesofbacterialoperationaltaxonomicunits(OTUs)withinandbetweenAe.aegyptilarvae,mosquitopositiveandmosquitonegative watersamples.Bracketsshowoutcomesoftwo-samplet-testcomparisonsofunweightedUniFracdistances;**P<0.01;ns,notsignificant. used to test the strength and significance of sample clus- Figure 5 shows the number of bacterial taxa obtained tering, showed that 20% of the variation explained by from TTGE sequences. A total of 24 taxa were identified PCoA was statistically significant (R2=0.20, p<0.001). overall,sevenwereidentifiedinlarvalsamples,16inmos- This indicates that the largest amount of variation, ex- quito positive and 13 in mosquito negative water samples plainedbyPCo1(Figure3),isstatisticallysignificant. (Figures4and5).Eightofthesetaxaweresharedbetween Comparing sequences from cultured water samples, mosquito positive and negative water samples (Figure 5), there was also no significant difference in OTU diversity while only two taxa, unclassified Bacilli and Actinomycet- between mosquito positive and mosquito negative sam- ales, were common to Ae. aegypti larvae and both mos- ples(p>0.05). quito positive and negative water samples (Figures 4 and 5). Otherthanthetwo bacterialtaxa common toall three BacterialtaxaassociatedwithTTGEsequences samples, no other taxon was shared between Ae. aegypti Four bacterial phyla and one candidate phylum (TM7) - a larvaeandwaterfromeithermosquitopositiveornegative majorlineageof bacteriaforwhichnoculturedrepresenta- containers (Figure 5). Unclassified Bacilli (26%) were the tives have been found, but whose existence is known from mostabundantbacterialtaxa in Ae. aegypti larvae, closely environmental 16SrRNA sequences - wereidentifiedfrom followed by unclassified Actinomycetales (25%) and Clos- Ae. aegypti larvae and water from domestic water con- tridium(11%).Theremaining38%comprisedunclassified tainers (Figure 4). Only two of these phyla, Actinobacteria Clostridiales (8%), Brevibacillus (4%), unclassified Bacilla- and Firmicutes, were found across all samples. The most ceae (1%), unclassified Firmicutes (1%), other bacterial abundant phylum identified from Ae. aegypti samples, was genera(19%)andunclassifiedsequences(5%).Unclassified Firmicutes (52%), followed by Actinobacteria (25%). Other Comamonadaceae(15%)werethemostabundantinmos- bacteria phyla (not broken down in QIIME) made up 18% quito positive water samples, followed by Acinetobacter of the mosquito samples and the remaining 5% were un- (13%), unclassified Proteobacteria (13%), unclassified classified sequences. Proteobacteria was the predominant Microbacteraceae (9%), Flavobacterium (9%), and unclas- phyluminbothmosquitopositiveandnegativewatersam- sified Actinomycetales (4%). The remaining 37% com- ples.Itconstituted50%ofbacteriaphylafoundinmosquito prisedotherbacterialgenera(20%),unclassifiedsequences positive water samples, followed by Actinobacteria (13%), (4%) and other small bacterial taxa contributing 1-2% of Bacteroidetes(9%),Firmicutes(2%),TM7(2%),otherphyla total bacteria genera (Figure 4). Similar groups of bac- (20%), and unclassified sequences (4%). In the mosquito teria dominated mosquito negative water samples, with negativewatersamples,Proteobacteriamadeup33%ofthe Flavobacterium (17%) being the most dominant, followed totalbacteriaphyla, followedbyActinobacteria(18%),Bac- by unclassified Comamonadaceae (10%), unclassified teroidetes (18%), Firmicutes (2%), TM7 (1%), other phyla Microbacteraceae (10%), unclassified Actinomycetales (21%)andunclassifiedsequences(7%). (8%), unclassified Sphingomonadales (8%), unclassified Dadaetal.Parasites&Vectors2014,7:391 Page7of12 http://www.parasitesandvectors.com/content/7/1/391 Figure3PCoAofunweightedUniFracdistancesbetweensamplesanalyzedbyTTGE.PrincipalcoordinateanalysisofunweightedUniFrac distancesbetweenAe.aegyptilarvaeandwaterfrommosquitopositiveandnegativecontainers.Distinctclustersoflarvalsamples(red)and mosquitopositivewatersamples(blue)arecapturedonallthreePCoAplots.Mosquitonegativewatersamples(orange)donotshowany distinctclustersonanyoftheplots.Eachaxisshowpercentageofvariationexplained.Eachdatapointconsistsofacentralpointsurroundedby ellipsoidsthatindicatevariationinUniFracdistancesfromrarefaction.Thisdemonstratesthattheclusteringpatternholdsuptosubsampling. Dadaetal.Parasites&Vectors2014,7:391 Page8of12 http://www.parasitesandvectors.com/content/7/1/391 Figure4RelativeabundanceofbacterialtaxaobtainedfromTTGEsequences.Barsshowmeanrelativeabundanceofthedifferentbacterial taxaisolatedbyTTGEfromAe.aegyptilarvae,mosquitopositiveandmosquitonegativewatersamples.**Sequencesthatwerenotclassifiedby Qiime.***LowabundancephylathatareautomaticallygroupedtogetherbyQIIME. Proteobacteria(6%),andRhodobacter(5%).Theremaining theidentifiedtaxa.Theother15%(n=2)comprisedmem- 36% comprised other bacterial taxa (21%), unclassified bersofthe classBetaproteobacteriaandwereonlyisolated sequences(7%),andsmallbacterialtaxacontributing1-2% from mosquito positive water samples (Figure 6). Four of oftotalbacterialtaxa(Figure4). theidentifiedbacterialtaxawereexclusivelyfrommosquito positive water samples, three from mosquito negative Bacterialtaxaassociatedwithsequencesfromcultured watersamples,andsixwere commontoboth.Acinetobac- watersamples ter was the most common bacterial genera isolated from Thirteen bacterial taxa belonging to the phylum Proteo- mosquitopositivewatersamples.Itconstituted37%ofthe bacteriawere identifiedfrom both mosquito positive and identified bacterial taxa, followed by Pseudomonas (20%), negative water samples (Figure 6). Bacteria belonging to Comamonas (13%), and unclassified Enterobacteriaceae the class Gammaproteobacteria made up 85% (n=11) of (12%). The remaining 18% comprised small bacterial taxa, Dadaetal.Parasites&Vectors2014,7:391 Page9of12 http://www.parasitesandvectors.com/content/7/1/391 Aedes aegypti larvae 7 2 16 13 8 Mosquitopositive Mosquitonegative watersamples water samples Figure5NumberofbacterialtaxaobtainedfromTTGE sequences.Venndiagramshowingnumberofbacterialtaxa obtainedfromTTGEanalysisofAe.aegyptilarvae,mosquitopositive andmosquitonegativewatersamples. including Escherichia/Shigella, making up between 0.8 – 5%ofidentifiedbacterialtaxa(Figure6).Acinetobacteralso dominated mosquito negative water samples, making up 27% of identified bacterial taxa. This was followed by unclassified Enterobacteriaceae (25%), Stenotrophomonas (20%) and Escherichia/Shigella (9%). The remaining 19% was made up of unclassified sequences (7%), and small bacterial genera contributing 1-4% of identified bacterialtaxa. Discussion Very sparse information is available on the nature of the microbial community associated with Ae. aegypti larvae Figure6Relativeabundanceofbacterialtaxaobtainedfrom indomesticwatercontainersinandaroundhumandwell- culturedwatersamples.Barsrepresentmeanrelativeabundance ofbacterialtaxaisolatedfrommosquitopositiveandnegativewater ings. Following our previous study [19], we hypothesized samplesculturedondrigalskiagar. that the bacteria in Ae. aegyptiinfested containersare de- terminantsofAe.aegyptiproductionandthusmayconsti- tute a major proportion of the larval microbiota. To test taxawithinmosquitolarvaeareyettobedescribed,orthat this,weutilizedthe16SrRNA-TTGEtocomparativelyas- methodsusedforscreeningarenotsufficienttoobtainthe sessthebacterialcommunitiesassociatedwithAe.aegypti whole picture of larval microbiota. While larvae consume larvae and water from these containers. In addition to bacteria,othermicrobessuchasalgaeandfungimaycon- TTGE, water samples were cultured for enteric bacteria. tributemoresignificantlytotheirdiet[33,34],whichcould This, to the best of our knowledge, is the first study beanotherreasonforlowbacterialOTUabundance. to compare bacterial communities associated with Ae. Although the impact of mosquito larvae on bacterial aegyptilarvaeandwaterfromdomesticcontainers. abundance and diversity in laboratory and field micro- Aedes aegypti larvae showed significantly lower OTU cosms have been variable [35], most studies report that abundance (Figures 1 and 2) compared to water samples the presence of larvae affects the bacterial diversity in fromdomesticcontainers.Thislowabundanceofbacterial breeding habitats [36-38]. Our study showed significantly taxa in mosquitoes compared to their breeding habitats different bacterial diversity between larvae and water has been frequently reported [5,31,32], indicating that samples (Figures 2, 3, and 4), with little overlap between mosquito larvae naturally have a low number of bacterial the bacterial communities. This may be because bacteria taxa. It may also mean that the majority of the bacterial already present in the larvae dominate and control the Dadaetal.Parasites&Vectors2014,7:391 Page10of12 http://www.parasitesandvectors.com/content/7/1/391 larval microbiota, thereby inhibiting establishment of new (Comamonadaceae, Polynucleobacter, Rubirivivax, and bacteria. Factors such as developmental stage, tissue trop- unclassified betaproteobacteria) and Actinobacteria ism, genetics, dynamics of intra- and inter-specific interac- (Actinomycetales, and Microbacteriaceae), were the tions, as well as environmental factors are thought to most abundant bacterial classes in mosquito positive influence the bacterial diversity within mosquitoes [10]. In water samples, with Gammaproteobacteria dominat- humans and other mammals, gut microbiota play an im- ing. Flavobacteria (Flavobacterium), Alphaproteobac- portant role in ‘colony resistance’, where they prevent teria (Rhodobacter, unclassified Sphingomonadales, colonization by other bacteria or pathogens [9]. There is and unclassified Rhizobiales), and Betaproteobacteria someevidenceofthis‘colonyresistance’ininsects[9,10],as (unclassified Comaonadaceae, unclassified Betaproteo- mosquito gut bacteria can either support or suppress the bacteri and Polynucleobacter) were the most abundant growth of other species by producing inhibitory factors inmosquitonegativewatersamples,withFlavobacteria [39]. Bacteria free in the mosquito gut lumen might evoke dominating. Enteric bacteria belonging to the class a host immune-defense response, or modify the gut envir- Gammaproteobacteria were isolated from both mos- onmentto inhibit development ofother bacteria[10,40]. It quito positive and negative water samples by selective is also possible that bacteria taken up by late larval instars bacterial culture (Figure 6). This class constituted 15% from their breeding containers are unable to colonize the of the total bacterial population isolated from mos- mosquitoesatthisstageoflarvaldevelopment.Thisispos- quito positive water samples using TTGE, but was not sibleduetotheselective,competitiveorprotectivemecha- identified in mosquito negative samples (Figure 4). It nisms elicited by established bacteria that may have been may have constituted the ‘other bacterial phyla’ not acquired at early stages of development. Internal competi- broken down in QIIME due to negligible proportions. tion among bacterial species could also be an explanation, This indicates that the concentration of Gammaproteo- as predominant bacterial taxa in mosquitoes may have bacteria may have been below TTGE detection limit, somecompetitiveadvantageoverothertaxa[41]. which is still debatable [18], or outweighed by high con- Overall, four bacterial phyla and one candidate phylum centrationsofothercompetingbacterialDNA[51]. (TM7) were identified from Ae. aegypti larvae and water Some bacteria are known to pass through 0.45 μm samples (Figure 4). Only two of these phyla, Firmicutes pore size membrane filters impacting bacterial density and Actinobacteria, were associated with both Ae. aegypti and, to a lesser extent, diversity. Hence some bacteria in larvae and water from their breeding containers, suggest- this study may havebeen lost during filtration.Nonethe- ingpossibledominanceandcontroloflarvalbacterialflora less, our results are in line with those of similar studies by these two phyla. The majority of the bacteria isolated conductedpreviously.Studiesonthebacterialcomposition from Ae. aegypti larvae belonged to the classes Bacilli of water samples from domestic containers have under- (Bacillaceae, Brevibacillus & unclassified bacilli), Acti- standably focused on fecal contamination [19,52-54], pro- nobacteria(Microbacteriaceae&unclassifiedActinomy- viding little information on general bacterial composition. cetales) and Clostridia (Clostridium & unclassified In theone study conducted so far, majority of the bacteria Clostridiales), with Bacilli being the most predominant. isolatedfromAe.aegyptiinfesteddomesticcontainerswere This is consistent with findings from other studies Proteobacteria, predominantly Gammaproteobacteria where bacteria belonging to these classes have been [9] which is in line with results reported here. In other identified in different mosquito species [15,42-45] in- mosquito habitats such as tree holes, tyres, discarded cludingAe.aegypti[41,46-50].Onestudyonthemidgut containers, plant pots, and laboratory mesocosms, Pro- microbiota of Ae. aegypti larvae collected from natural teobacteria have also been shown to be predominant breeding habitats in Thailand identified Bacilli (Bacillus [5,9,44]. Isolation of enteric bacteria from Ae. aegypti cereus) as the most predominant bacterial class [48]. In infested domestic water containers supports findings contrast, Gammaproteobacteria was the most abundant from our earlier study [19] where these containers were class (64%) in Ae. aegypti larvae collected from domes- more likely to be contaminated with E. coli (an enteric tic containers in Pune and Ahemedabad, India [9]. The bacteria) than not. This suggests that enteric bacteria TTGE results in this study did not identify bacteria mayplayaroleinAe.aegyptiinfestationinthissetting. from this class or phylum in Ae. aegypti larvae. This maybeduetodifferencesinlocationortechniquesused Conclusions forisolatingbacteria,orboth. We present for the firsttime results ofthe bacterialcom- Proteobacteria was the most predominant phylum positionoffourth-instarAe.aegyptilarvaeandwaterfrom in both mosquito positive and negative water samples. Ae. aegypti infested domestic water containers. Aedes The TTGE results showed that Gammaproteobacteria aegyptihadsignificantlylowerOTUabundancecompared (Acinetobacter, unclassified Gammaproteobacteria, to mosquito positive water samples. There was no signifi- and unclassified Legionellaceae), Betaproteobacteria cant difference in OTU abundance between larvae and
Description: